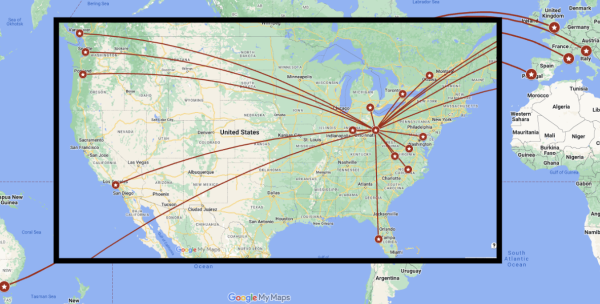
The Department of Statistics faculty and students have been globe-trotting on a professional mission once again this year. While travel had slowed during the global pandemic, our faculty and students are back to meeting colleagues around the country and world. The graphic above visualizes statistics and biostatistics graduate student travel over 2023-24, including 52 trips for 36 unique students to 22 unique events. We are extremely proud of the students who travel to professional events on their own without the ready-made safety net of close colleagues. We are also proud of those who confront the sometimes overwhelming crowd and buzz of activity at very large events to present their work. Across the board, we are proud of our students’ work and are glad that they are making an impact on their colleagues and the future of our field by sharing it openly.

The Joint Statistical Meetings are easily the most well-attended for both students and faculty in the department – and also alumni and good friends. This year, many department affiliates and friends joined us at the Joint Statistical Meetings in Portland as we co-hosted — along with the Division of Biostatistics — The Ohio State Statistics and Biostatistics Mixer! This marked the kick-off for our 50th Anniversary celebration. The Mixer took place at Screen Door on August 4, 2024 and featured fantastic food and conversation. Several student attendees commented on the gap in food quality between the Mixer and our usual post-seminar reception!
Make plans now to join us at next year's mixer in Nashville during JSM 2025! While we won’t have such a momentous reason to celebrate, we still intend to have a great event in the evening of Sunday, August 3. With the proximity to Columbus, we expect many department affiliates and friends will attend.
While fellowship with colleagues and friends is important, the meat of the conference is the research! Below is a list of work presented at JSM 2024 by department faculty and their students.
Graduate student travel is made possible in part by the generosity of those who donate to the Gary G. Koch and Family Graduate Student Travel Award Endowment Fund.
Steephanson Anthonymuthu (Assistant Professor of Teaching Practice) presented “A latent variable approach for the estimation of the mitigated fraction for ordinal response” on behalf of his co-author Christopher Vahl
The efficacy of an intervention, such as a vaccine, can be established through the estimation of several numerical measures. Mitigated fraction is one of the contemporary numerical measures, and it serves the purpose of reducing the severity of a specific disease rather than completely preventing its occurrence. Here, a novel approach to calculating the mitigated fraction is presented, which involves utilizing the values of latent variables within a generalized linear mixed model (GLMM). The proposed method addresses a pressing need in the field, particularly given the increasing interest in assessing efficacy measures for ordinal responses, especially when observations are clustered or measured longitudinally. The USDA's Center for Veterinary Biologics (CVB) recommends a form of the mitigated fraction which can be calculated when the disease severity can be graded by some continuous measure or by some discrete assessment resulting in unambiguous ranks. Results show the proposed latent variable approach works well for data collected in a completely randomized design (CRD) and a randomized complete block design (RCBD).
Yingyu Cheng (PhD Student, Statistics) presented “Bayesian Calibration of Material Properties for Shape Memory Alloys from Nanoindentation Data” on behalf of her co-authors Oksana Chkrebtii (Associate Professor), Peter Anderson, Daniel Hong, Steve Niezgoda and Thomas Santner (Professor Emeritus)
Shape memory alloys (SMAs) are materials with exceptionally high recoverable strains and temperature-sensitive material response, seeing growing use in medical and aerospace applications. We contribute to the development and quality control of SMAs by combining mathematical models with physical testing data to understand their mechanical properties. Nanoindentation is an experimental method where a shaped tip indenter is pushed into a material surface while recording applied force and penetration depth, providing information about material deformation. We study the problem of inferring thermomechanical properties of SMAs given experimentally obtained indentation curves at nano-micron scales. Sampling and spatial variation in the local grain structure at each indentation site result in spatially-varying material properties modeled as random effects, while the computationally expensive constitutive model is learned from a training sample of finite element simulations within a Bayesian hierarchical model. Bayesian calibration characterizes SMA material properties with associated uncertainty based on data from adaptively selected indentation locations and multimodal tip shapes.
Ye Jin Choi (PhD Student, Biostatistics) presented “Analyzing Spatial Dependence in Functional Data and Shapes of 2D Curves” on behalf of his co-authors Karthik Bharath and Sebastian Kurtek (Professor)
In this work, we model the shapes of spatially dependent functional data or boundaries of two-dimensional (2D) objects, i.e., spatially dependent shapes of parameterized curves. Functional data is often composed of two confounded sources of variation: amplitude and phase. Amplitude captures shape differences among functions while phase captures timing differences in these shape features. Similarly, boundaries of 2D objects represented as parameterized curves exhibit variation in terms of their shape, translation, scale, orientation and parameterization. We study the spatial dependence among functions or curves by first decomposing given data into the different sources of variation. The proposed framework leverages a modified definition of the trace-variogram, which is commonly used to capture spatial dependence in functional data. We propose different types of trace-variograms that capture different components of variation in functional or shape data, and use them to define a functional/shape mark-weighted K function by considering their locations in the spatial domain as random. This statistical summary then allows us to study the spatial dependence in each source of variation separately. Efficacy of the proposed framework is demonstrated through extensive simulation studies and real data applications.
Wenxin Du (PhD Student, Statistics) presented “Space-Time Bayesian Modeling of Virtual Reality Behavioral Data” on behalf of her co-authors Mario Peruggia (Professor), Deborah Kunkel (PhD 2018, Statistics), Shadrick Addy, and Hsi-Yuan Chu
Behavioral data in Virtual Reality (VR) provides insight into understanding how users interact and shift their focus of attention within a virtual environment. The user-centric nature of VR experiences, emphasizing a first-person perspective, allows the representation of informative data (such as head orientation and rotation) as directional data, which can be mapped onto a sphere in the 3-dimensional space. Postulating that eye direction is a representation of VR users' focus of attention, we designed a VR experiment to study how head behavioral data collected continuously through the VR headset worn by participants can be used to estimate their focus of attention. For data analysis, we construct hierarchical Bayesian models using the Kent distribution (also known as the 5-parameter Fisher–Bingham distribution). Throughout, we propose a focus tracking method based solely on head behavioral data that may serve as a proxy to eye-tracking technology, which gives accurate representation of focus but relies on expensive equipment and raises privacy and safety concerns.
Alan Gan (PhD Student, Statistics) presented “A Bayesian perspective to finite element methods for time-dependent partial differential equations” on behalf of his co-author Oksana Chkrebtii (Associate Professor)
Partial differential equations (PDEs) are crucial in modeling physical phenomena, capturing complex dynamics such as heat transfer and fluid flow. Numerical solvers approximate solutions of PDEs through discretization, thereby introducing error and uncertainty in these numerical solutions. There has been recent interest and work in modeling and quantifying this error probabilistically, allowing to more comprehensively account for sources of error when, for instance, solving a Bayesian inverse problem. To that end, we present a Bayesian probabilistic method to model the spatial and temporal discretization error in finite element method (FEM) solvers for time-dependent PDEs. We apply our method on linear and nonlinear PDE test problems, and integrate it into a Bayesian inverse problem.
Chris Hans (Associate Professor) presented “Marginal likelihood estimation for Bayesian lasso-like regression” on behalf of his co-author Ningyi Liu (PhD Student, Statistics)
Formal Bayesian methods for model comparison depend on the marginal likelihood of the data given a model. When closed-form expressions for marginal likelihoods in a given model class are not available, it is common to employ computational approaches to either estimate the marginal likelihoods directly or to avoid their explicit evaluation by summarizing output from carefully constructed MCMC algorithms. While Bayesian treatments of approaches to penalized regression have become popular tools for data analysis, formal Bayesian model comparison in these settings can be challenging. Computing marginal likelihoods for Bayesian lasso-like regression models is difficult due to the L1-norm penalty term that is incorporated into the prior on the regression coefficients. We introduce MCMC-based approaches for estimating the marginal likelihoods for the Bayesian lasso and elastic net regression models. The methods involve sampling from standard probability distributions, need not rely on any data augmentation, and require no tuning of random walks or specification of approximating distributions. We make comparisons to other related approaches for marginal likelihood estimation.
Zhenbang Jiao (PhD Student, Statistics) presented “When cross-validation meets Cook's distance” on behalf of his co-author Yoonkyung Lee (Professor)
We introduce a new feature selection method for regression models based on cross-validation (CV) and Cook's distance (CD). Leave-one-out (LOO) CV measures the difference of the LOO fitted values from the observed responses while CD measures their difference from the full data fitted values. CV selects a model based on its prediction accuracy and tends to select overfitting models often. To improve CV, we take into account model robustness using CD, which can be shown to be effective in differentiating overfitting models. Hence we propose a linear combination of CV error and the average Cook's distance as a feature selection criterion. Under mild assumptions, we show that the probability of this criterion selecting the true model in linear regression using the least squares method converges to 1, which is not the case for CV. Our simulation studies also demonstrate that this criterion yields significantly better performance in feature selection for both linear regression and penalized linear regression compared to CV. As for computational efficiency, this criterion requires no extra calculation compared to CV as CD involves the same fitted values needed for CV.
Yoonkyung Lee (Professor) presented “Measuring case influence locally”
In statistical modeling, assessing case influence is important for model diagnostics. It does not only help us identify unusual observations for further investigation and model refinement, but it also allows us to understand model sensitivity. While case deletion is the most common form of data perturbation for studying case influence, it is computationally expensive in general as it requires refitting models. Alternatively, we consider model sensitivity to an infinitesimal data perturbation known as local influence, and show that this notion of local influence is broadly applicable to models in regression and classification. We use a case influence graph as a tool for assessing influence with a continuous case weight and examine the impact of a small perturbation in the case weight on the model to measure local influence. Once the case-weight adjusted model is identified, measures of local influence can be calculated easily using the full data model alone. We demonstrate this approach through applications to quantile regression and support vector machines.
Shili Lin (Professor) participated in the panel discussion “Florence Nightingale Day Inspires Students to Pursue Statistics and Data Science” along with fellow panelists Donald Estep, Jessica Kohlschmidt (PhD 2009, Statistics), Sideketa Fofana, and David Kline (PhD 2015, Biostatistics)
Florence Nightingale was a pioneer in data visualization and the founder of modern nursing. She used data in a novel and effective way to provide better care for wounded soldiers in the Crimean War and for public health improvements more broadly. Her pioneering work included the creation of the pie chart, which is universally used in data visualization to this day. To honor Nightingale's legacy, the Florence Nightingale Day (FN Day) was jointly launched by ASA and CWS (Caucus for Women in Statistics) in 2018 to engage middle and high school students, promote future career opportunities in statistics and data science, and celebrate the contributions of women to these fields. The vision is to inspire a diverse group of young people to become future leaders in data-related disciplines. Pre-college students (ages 13 and above) from around the world are encouraged to attend this one-day free event. Components of the day include trivia games, panels of professional and student speakers, hands-on activities, and networking. In this session, the panelists will discuss their experiences hosting their FN Day in the United States and Canada. We will feature panelists whose sites have successfully recruited women and students from disadvantaged groups, and those from institutions that enroll over 85% of Hispanic students. Our panelists will also share information on how to host your own FN Day.
Yue Ma (PhD Student, Statistics) presented “Gaussian processes with continuous boundary restrictions: physical constraints for spatial models” on behalf of co-authors Oksana Chkrebtii (Associate Professor) and Steve Niezgoda
Boundary constraints are used extensively in engineering and physical models to restrict continuous states to follow known physical laws. Examples include fixed-state or fixed-derivative (insulated) boundaries, and boundaries which relate the state and the derivative (e.g., radiative or absorbing boundaries). We develop a flexible framework to construct Gaussian processes which fully enforce linear boundary constraints on simply-connected domains. This flexible class of stochastic processes can be used for recovering a smooth field, such as material stress, from discrete spatial measurements across a domain with known physical mechanisms working at the domain boundaries. Such processes make flexible priors for modeling the states of dynamical systems defined by partial differential equations (PDEs), have applications in constructing probabilistic numerical solvers, and enable data-driven discovery of PDEs. We generalize our approach to a wide range of covariances such as Matérn, and various domains such as disks. As examples, we simulate heat flow in the Jominy End Quench test, recover Burger's equation from data, and infer a stress field across a material sample.
Jillian Morrison (Assistant Professor of Teaching Practice) presented “Performance Comparison of Models for Analysis of Ordinal Data” on behalf of her co-authors Matthew Jobrack and Nairanjana Dasgupta
Motivated by a real data, this is a "what if" simulation paper: where we knowingly use a theoretically incorrect model to see what exactly the penalties are. The consequences of using Ordinary Least Squares (OLS) when the response is ordinal in nature are explored. Specifically, whether Type I error is maintained and if there is a loss in power for OLS when looking at the relevance of various explanatory variables in comparison to the "correct'' Generalized Linear Models (GLM) are addressed. Results show that when looking at relevance of explanatory variables in the context of tests for slopes, there is no appreciable loss in power. Further, Type I error is well maintained using OLS methods where the ordinal logit often results in inflated Type I errors.
Subhadeep Paul (Associate Professor) presented “Measurement error and network homophily in autoregressive models of peer effects”
The autoregressive models of peer effects include the SAR model in cross sectional studies used to estimate the peer influence and the effects of covariates taking network dependence into account, and the longitudinal model to causally identify the effect of peer actions in the preceding time period. We investigate issues of measurement error and network homophily in both of these setups.
First, we investigate causal peer role model effect on successful graduation from Therapeutic Communities (TCs) for substance abuse using records of exchanges among residents and their entry and exit dates which allowed us to form peer networks and define a causal estimand. To identify peer influence in the presence of unobserved homophily, we model the network with a latent variable model and show that our peer influence estimator is asymptotically unbiased. Second, in the context of SAR model, while the model can be estimated with a QMLE approach, the detrimental effect of covariate measurement error on the QMLE and how to remedy it is currently unknown. We develop a measurement error-corrected ML estimator and show that it possesses statistical consistency and asymptotic normality properties.
Mario Peruggia (Professor) presented “Fingerprinting Bayesian Gaussian Mixture Models” on behalf of his co-author Wenxin Du (PhD Student, Statistics)
Case deletion analysis enables one to quantify the impact that individual observations (or groups of observations) exert on the posterior distribution when they are included in the analysis. For each observation, its case deletion weight is defined as the ratio of the case-deleted posterior to the full posterior. The covariance matrix of the log case deletion weights is rich with information about the fit of the model and can be used to uncover possible sources of lack of fit. In this talk, we study the behavior of the eigenvalues of this matrix, and we show how their properties can be used to learn about how many components should be specified in a Bayesian Gaussian mixture model. In simpler situations we can establish exact distributional results. In more complex situations, we validate our results through extensive simulation studies.
Sean Tomlin (PhD Student, Biostatistics) presented “Matched difference-in-differences design with counterparts”
Difference-in-differences (DiD) is a widely used design for evaluating the impact of interventions on health outcomes. DiD designs often use parametric models by assuming the untreated potential outcomes evolve similarly over time. However, this parallel trends assumption may be marginally violated, which can bias the estimate of the average treatment effect on the treated (ATT). An alternative assumption is conditional parallel trends, which posits parallel trends within strata. Matching can facilitate conditional parallel trends in a DiD design. However, mechanical application of matching techniques to DiD designs may facilitate conditional ignorability but not parallel trends. This stems from a flawed analogy: controls closely resemble treated individuals except for the treatment. Instead, this matched design contains counterparts (a type of nonequivalent control) that are similar in overt ways (covariates) but different in hidden ways that can be well-defined (time trend). With this reinforced analogy, we clarify the assumptions of matched DiD studies and the role of counterparts in design & analysis, and we revisit the regression to the mean problem in inference about the ATT.
Emerson Webb (PhD Student, Statistics) presented “A Divide and Conquer Strategy for Recapitulating Whole Genome 3D Structure Using Hi-C Data" on behalf of first author Jincheol Park and co-authors Meng Wang and Shili Lin (Professor).
The 3D organization of the genome is closely linked to biological functions. High throughput assays such as bulk Hi-C capture genome-wide chromatin interactions; such information can be used to discern the 3D organization. Methods for inferring 3D structures from Hi-C data are either optimization- or sampling-based. Optimization-based methods can construct whole genome 3D structure but do not account for cell heterogeneity in bulk data. Sampling-based methods generate a distribution of possible 3D structures to account for cell heterogeneity. However, whole-genome 3D structure recapitulation is too computationally expensive for sampling-based methods; therefore, chromosome-by-chromosome strategies are usually used, but doing so ignores important information on inter-chromosomal contacts. We overcame these issues by proposing the truncated Random effect EXpression-Cut And Paste (tREX-CAP) method, which uses the tREX model coupled with a divide-and-conquer strategy to efficiently infer the whole genome 3D structure. We evaluated the performance of tREX-CAP and compared to two other methods through an extensive simulation study and an analysis of a bulk Hi-C lymphoblastoid dataset.
Paul Wiemann (Assistant Professor) presented “Scalable non-Gaussian VI for continuous functions using sparse autoregressive normalizing flows” on behalf of his co-author Matthias Katzfuss
We introduce a novel framework for scalable and flexible variational inference targeting the non-Gaussian posterior of a latent continuous function or field. For both the prior and variational family, we consider sparse autoregressive structure corresponding to nearest-neighbor directed acyclic graphs, where the conditional distributions in the variational family are modeled using highly flexible normalizing flows. We provide an algorithm for doubly stochastic variational optimization, achieving logarithmic time complexity per iteration. We present numerical comparisons that illustrate the proposed method can be more accurate than a Gaussian variational family while maintaining the same computational complexity.
Biqing Yang (PhD Student, Statistics) presented “A Bayesian Approach for Achieving Double Robustness in Treatment Effect Estimation” on behalf of her co-author Xinyi Xu (Professor)
Combining propensity and prognostic scores enhances the efficiency of matching methods in estimating average treatment effect in observational studies. This paper aims to provide a Bayesian approach of double score estimation as well as a theoretical support of the consistency of the Bayesian estimator. Specifically, we explore the performance of a semiparametric Bayesian model, utilizing Gaussian process priors and addressing potential model mis-specification. We derive asymptotic results to validate the consistency of Bayesian estimators as the sample size increases. Particularly noteworthy is the demonstrated superiority of double-score Bayesian estimators in estimating both the population and conditional average treatment effects. In the simulation study, we analyze the performance of these models under various scenarios with a finite sample size. The results generated by the MCMC algorithm indicate doubly robust estimation under specific conditions. We also apply our proposed single/double score model to a real-world dataset, yielding results that align with existing studies utilizing matching methods.
Haozhen Yu (PhD Student, Statistics) presented “Case Sensitivity in Regression and Beyond” on behalf of her co-author Yoonkyung Lee (Professor)
The sensitivity of a model to data perturbations is key to model diagnostics and understanding model stability and complexity. Case deletion has been primarily considered for sensitivity analysis in linear regression, where the notions of leverage and residual are central to the influence of a case on the model. Instead of case deletion, we examine the change in the model due to an infinitesimal data perturbation, known as local influence, for various machine learning methods. This local influence analysis reveals a notable commonality in the form of case influence across different methods, allowing us to generalize the concepts of leverage and residual far beyond linear regression. At the same time, the results show differences in the mode of case influence, depending on the method. Through the lens of local influence, we provide a generalized and convergent perspective on case sensitivity in modeling that includes regularized regression, large margin classification, generalized linear models, and quantile regression.
Xinyu Zhang (PhD Student, Statistics) presented “Diagnostic measures for Bayesian restricted likelihood”
Bayesian restricted likelihood makes use of a formal model for the data to move from prior distribution to posterior distribution. The twist is that the Bayesian update is based on an insufficient statistic, typically chosen to enhance the robustness of conclusions to potential deficiencies in the formal model. Computational methods that rely on MCMC algorithms include a step where an artificial data set is generated that matches the observed value of the insufficient statistic. The generation of these artificial data sets provides access to a full suite of diagnostics, both graphical and numerical, that allow the analyst to assess the adequacy of the formal model. The diagnostics may suggest ways in which the formal model can be improved.
Zhizhen Zhao (PhD Student, Statistics) presented “M-learning for Individual Treatment Rule with Survival Outcomes” on behalf of her co-authors Andi Ni, Xinyi Xu (Professor), Bo Lu, and Macarius Donneyong
Individualized treatment rules (ITRs) tailor treatments to individuals based on their unique characteristics to optimize clinical outcomes and resource allocation. In this paper, we expand the existing matched-learning (M-learning) methodology to estimate optimal ITRs under right censored data, as time-to-event outcomes are common in medical research. We construct matched sets for individuals by comparing observed times and incorporate an inverse probability censoring weight into the value function to handle censored observations. Additionally, we propose a full matching design in M-learning to reduce the potential overuse of a single subject in matching with replacement. We show that the proposed value function is unbiased for the true value function without censoring. We conduct a simulation study to compare the proposed method with the existing M-learning approach and a weighted learning approach. Results are evaluated based on winning probabilities and estimated values. Different methods show somewhat different performance under various scenarios. Finally, we apply the proposed method to estimate optimal ITRs for patients with Atrial fibrillation (AF) complications.
